Early diagnosis and quarantine of the coronavirus is an important step in curbing further spread.
Coronavirus disease (COVID 19) is an infectious disease caused by the coronavirus. This affects the respiratory system of the infected person and in most cases, they recover without any special treatment. It can turn into a serious illness for older people and those with underlying medical problems like cardiovascular disease, diabetes, chronic respiratory disease, and cancer.
With the pandemic in place, early diagnosis and prevention steps are necessary to curb the spread of the virus.
While preventive measures include wearing masks, social distancing, and frequent sanitization, it is also necessary to diagnose the presence of the disease at its early stages in order to quarantine the affected and prevent further spreading.
Deep learning models have proven useful and very efficient in the medical field to process scans, x-rays, and other medical data to output useful information.
In this article, we use lung CT (computed tomography) scans of people to diagnose the presence of coronavirus in their system.
A link to the implementation on cAInvas — here.
The dataset
The dataset folder consists of 397 non-COVID and 349 COVID lung CT scans. This seems to a fairly balanced dataset. A .xlsx file containing some metadata is also available, but we will not be using that here.
The images are collected from COVID19-related papers from medRxiv, bioRxiv, NEJM, JAMA, Lancet, etc., and available for use on Kaggle. (The links in the notebook mentioned above facilitate direct download and use of the dataset).
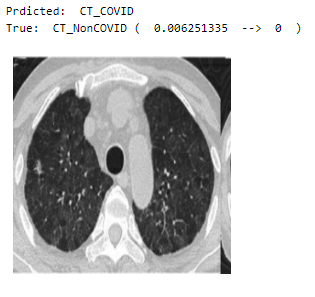
Here are a few images of both categories (CT_COVID and CT_NonCOVID) —
Each image is of size 256×256 with 3 channels.
The dataset is divided into train and test in an 80–20 ratio. Since the images are readily available as two folders, we use the image_dataset_from_directory function of the tensorflow.keras.preprocessing module to create train and validation sets by specifying the appropriate parameter values.

Preprocessing
The only preprocessing we will be doing is normalizing the pixel values.
We use the Rescaling function of keras.layers.preprocessing module to divide each value by 255. Thus the range of pixel values will now be [0, 1].
The model
The model has three pairs of Conv2D-MaxPooling2D layers followed by some Dense layers, all with ReLU activation except the final one, which has a sigmoid activation and 1 node in the dense layer.
The model is compiled using Adam optimizer with a learning rate of 0.001.
The BinaryCrossentropy loss function is used because the final dense layer has a sigmoid activation function. The accuracy of the model on the data is tracked.
The model is trained for 16 epochs.
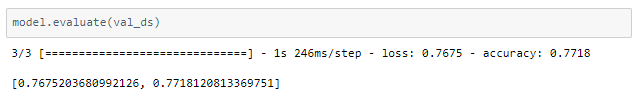
It seems to be a reasonable model but there is a difference in accuracy between the train and validation accuracy. This high variance can be reduced by training with a larger dataset, thus resulting in higher accuracy.
The above model cannot be directly applied to real-time data for diagnosis in hospitals due to the low accuracy. But this is a proof of concept that lung CT scans are promising in providing accurate, fast, and cheap screening and testing of COVID-19.
The metrics
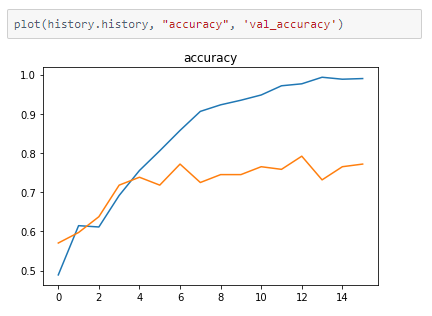
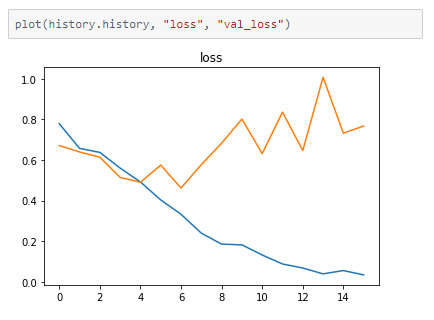
Prediction
Visualizing the predictions of random values in the validation set
deepC
deepC library, compiler, and inference framework are designed to enable and perform deep learning neural networks by focussing on features of small form-factor devices like micro-controllers, eFPGAs, CPUs, and other embedded devices like raspberry-pi, odroid, Arduino, SparkFun Edge, RISC-V, mobile phones, x86 and arm laptops among others.
Compiling the model with deepC to get .exe file —
Head over to the cAInvas platform (link to notebook given earlier) and check out the predictions by the .exe file!
Credit: Ayisha D
Also Read: Hate Speech and Offensive Language Detectionreplica rolex cellini
