Heart disease refers to any condition affecting the heart. There are many types, some of which are preventable. Share on Pinterest mikroman6/Getty Images. Unlike a cardiovascular disease, which includes problems with the entire circulatory system, heart disease affects only the heart.
This project will focus on predicting heart disease using neural networks. Based on attributes such as blood pressure, cholesterol levels, heart rate, and other characteristic attributes, patients will be classified according to varying degrees of coronary artery disease. This project will utilize a dataset of 303 patients and distributed by the UCI Deep Learning Repository.
We will be using some common Python libraries, such as pandas, NumPy, and matplotlib. Furthermore, for the deep learning side of this project, we will be using sklearn and Keras.
In this project, we are going into the sequence, and follow steps one by one:
Importing necessary libraries
Importing the Dataset
Now, we are importing the dataset or say we are reading the dataset.
This dataset contains patient data concerning heart disease diagnosis that was collected at several locations around the world. There are 76 attributes, including age, sex, resting blood pressure, cholesterol levels, echocardiogram data, exercise habits, and many others.
To data, all published studies using this data focus on a subset of 14 attributes — so we will do the same. More specifically, we will use the data collected at the Cleveland Clinic Foundation.
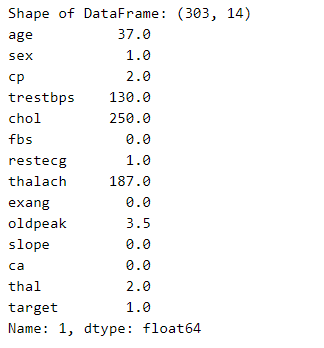
Now, we are printing the dataframe, so we can see how many examples we have.
Now, for preprocessing the data, we remove missing data (indicated with a “?”).
Now, we are dropping the rows with NaN values from DataFrame.
Now, we transform data to numeric to enable further analysis.

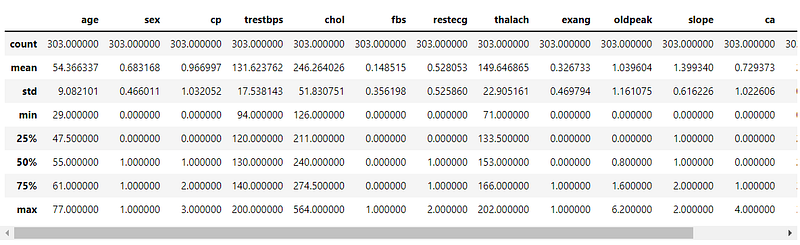
Now, we print data characteristics, using pandas built-in describe() function.
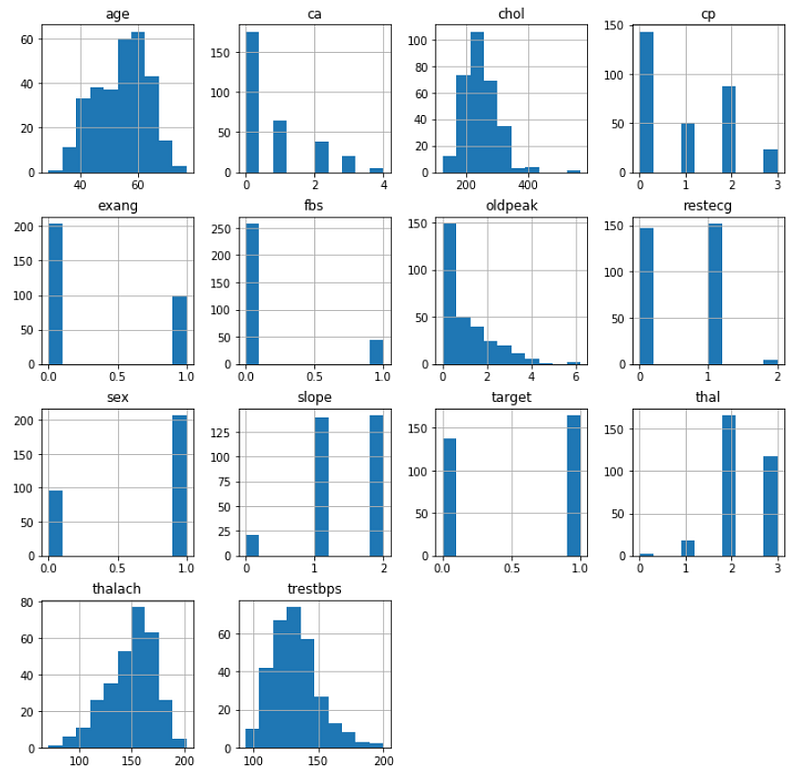
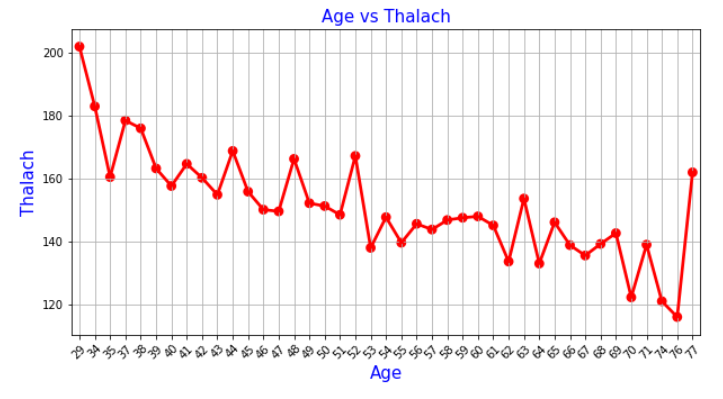
Now, we are plotting the histograms for each variable.
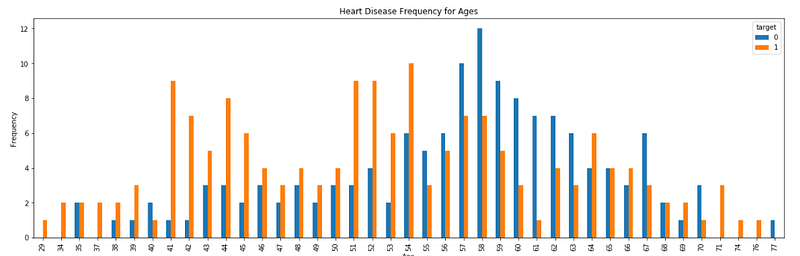
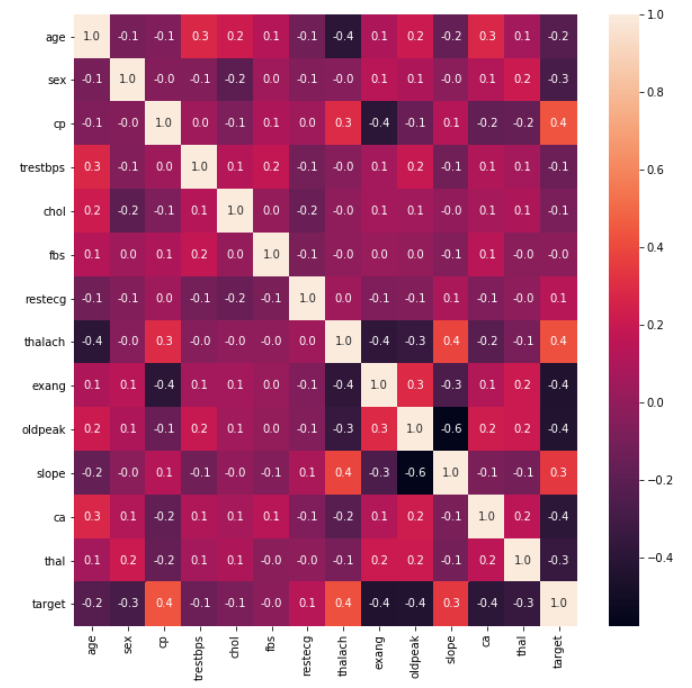
Create Training and Testing Datasets
Now that we have preprocessed the data appropriately, we can split it into training and testing datasets. We will use Sklearn’s train_test_split() function to generate a training dataset (80 percent of the total data) and a testing dataset (20 percent of the total data).
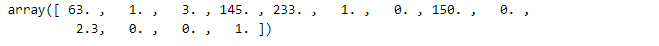

Now, we are creating X and Y datasets for training.
Then, we convert the data to categorical labels.
Building and Training the Neural Network
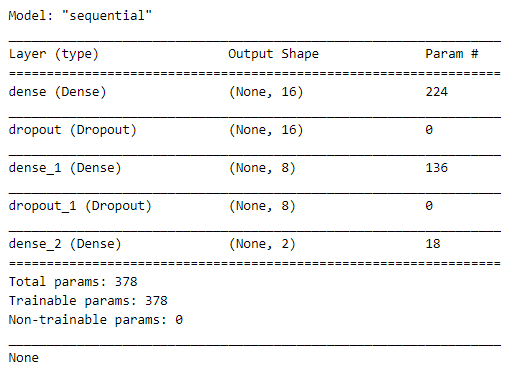
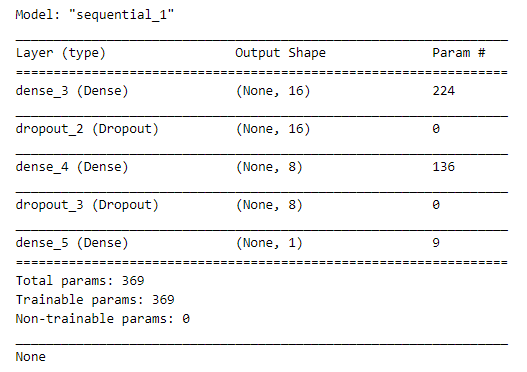
Now that we have our data fully processed and split into training and testing datasets, we can begin building a neural network to solve this classification problem. Using Keras, we will define a simple neural network with one hidden layer.
Since this is a categorical classification problem, we will use a softmax activation function in the final layer of our network and a categorical_crossentropy loss during our training phase.
Now, we fit the model to the training data.
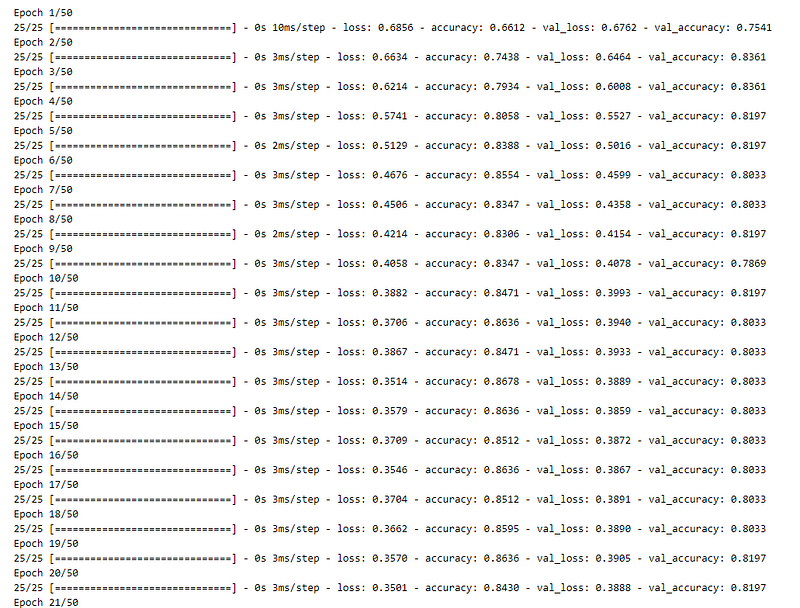
Now, we are plotting the graph of model accuracy.
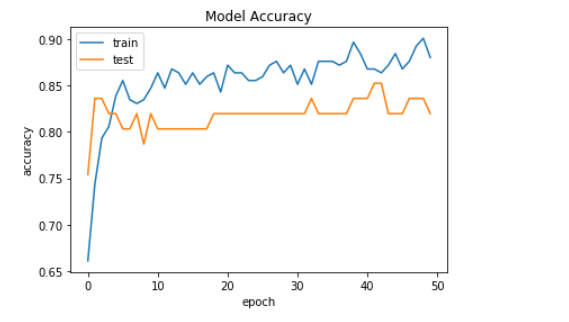
Now, we are plotting the graph of model loss.
Improving Results — A Binary Classification Problem
Although we achieved promising results, we still have a fairly large error. This could be because it is very difficult to distinguish between the different severity levels of heart disease (classes 1–4). Let’s simplify the problem by converting the data to a binary classification problem — heart disease or no heart disease.

Now, we define a new Keras model for binary classification, and then later we also check the model accuracy and model loss by plotting their required graphs.
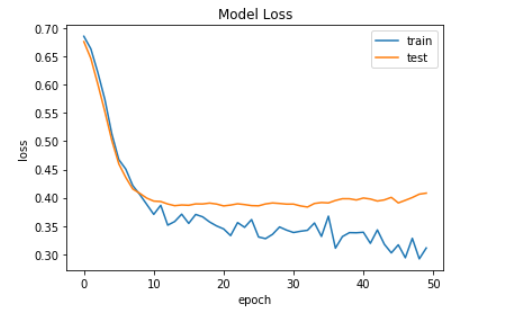
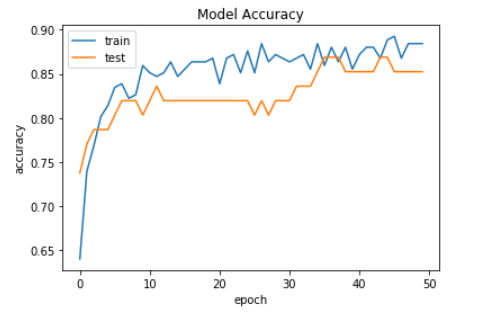
Now, we plot the graph of model accuracy but this time this is for the binary classification model.
Now, we plot the graph of model loss.

Results and Metrics
The accuracy results we have been seeing are for the training data, but what about the testing dataset? If our models cannot generalize to data that wasn’t used to train them, they won’t provide any utility.
Let’s test the performance of both our categorical model and binary model. To do this, we will make predictions on the training dataset and calculate performance metrics using Sklearn.
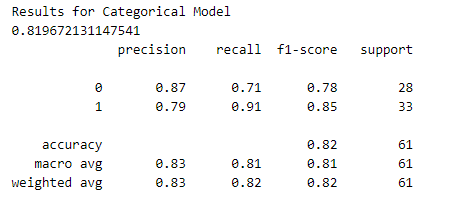
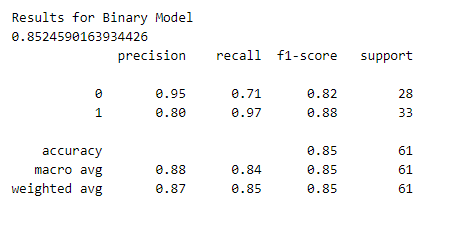
Also Read: Brain Tumor Detection
Now, we save our model
This is all about the heart disease prediction project.
You can download or go through the notebook from the link given here.
Credit: Bhupendra Singh Rathore
You may also be interested in
- Learning more about Parkinson’s Disease Detection using Spiral Drawings and CNN
- Reading about Resume Screening using Deep Learning on Cainvas
- Finding out about Neural network, the brain-inspired algorithms revolutionizing how machines see and understand the world around us
Become a Contributor: Write for AITS Publication Today! We’ll be happy to publish your latest article on data science, artificial intelligence, machine learning, deep learning, and other technology topics.
