Built a Deep Learning model using TensorFlow and Keras for grape leaf disease detection
Aim
To develop a Deep Learning model to identify diseased grape leaves using python programming language and Deep learning libraries
Prerequisites
Before getting started, you should have a good understanding of:
- Python programming language
- Deep Learning Libraries(Tensorflow, Keras, OpenCV)
Dataset
Link to download the dataset:
URL: https://cainvas-static.s3.amazonaws.com/media/user_data/vomchaithany/Grapes-Leaf-Disease-data.zip
get and unzip the data
output:
--2021-06-28 13:36:55-- https://cainvas-static.s3.amazonaws.com/media/user_data/vomchaithany/Grapes-Leaf-Disease-data.zip Resolving cainvas-static.s3.amazonaws.com (cainvas-static.s3.amazonaws.com)... 52.219.64.72 Connecting to cainvas-static.s3.amazonaws.com (cainvas-static.s3.amazonaws.com)|52.219.64.72|:443... connected. HTTP request sent, awaiting response... 200 OK Length: 23864365 (23M) [application/zip] Saving to: ‘Grapes-Leaf-Disease-data.zip’
Grapes-Leaf-Disease 100%[===================>] 22.76M 81.8MB/s in 0.3s
2021-06-28 13:36:55 (81.8 MB/s) - ‘Grapes-Leaf-Disease-data.zip’ saved [23864365/23864365]
Preparing data for the model
we assign train data to dirtrain and test data to dirtest
we are going to deal with 4 categories :
- Black_rot
- Esca_(Black_Measles)
- Health
- Leaf_blight_(Isariopsis_Leaf_Spot)
output:
Converting the images into arrays and storing them with labels
shuffling the data
Resizing the images
output:
Creating the pickle files
Model
output:
Model: “sequential”
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
conv2d (Conv2D) (None, 256, 256, 32) 896
_________________________________________________________________
conv2d_1 (Conv2D) (None, 254, 254, 32) 9248
_________________________________________________________________
max_pooling2d (MaxPooling2D) (None, 31, 31, 32) 0
_________________________________________________________________
conv2d_2 (Conv2D) (None, 31, 31, 32) 9248
_________________________________________________________________
conv2d_3 (Conv2D) (None, 29, 29, 32) 9248
_________________________________________________________________
max_pooling2d_1 (MaxPooling2 (None, 3, 3, 32) 0
_________________________________________________________________
activation (Activation) (None, 3, 3, 32) 0
_________________________________________________________________
flatten (Flatten) (None, 288) 0
_________________________________________________________________
dense (Dense) (None, 256) 73984
_________________________________________________________________
dense_1 (Dense) (None, 4) 1028
=================================================================
Total params: 103,652
Trainable params: 103,652
Non-trainable params: 0
_________________________________________________________________
output:
Epoch 1/10 2/42 [>.............................] - ETA: 4s - loss: 33.4274 - accuracy: 0.4062WARNING:tensorflow:Callbacks method `on_train_batch_end` is slow compared to the batch time (batch time: 0.0359s vs `on_train_batch_end` time: 0.0988s). Check your callbacks. 42/42 [==============================] - 6s 140ms/step - loss: 4.3365 - accuracy: 0.4440 - val_loss: 5.3007 - val_accuracy: 0.3276 Epoch 2/10 42/42 [==============================] - 6s 135ms/step - loss: 1.2223 - accuracy: 0.5103 - val_loss: 2.2340 - val_accuracy: 0.4095 Epoch 3/10 42/42 [==============================] - 6s 134ms/step - loss: 0.8702 - accuracy: 0.6398 - val_loss: 1.6332 - val_accuracy: 0.4698 Epoch 4/10 42/42 [==============================] - 5s 120ms/step - loss: 0.6871 - accuracy: 0.7380 - val_loss: 0.5808 - val_accuracy: 0.7198 Epoch 5/10 42/42 [==============================] - 6s 133ms/step - loss: 0.5256 - accuracy: 0.7761 - val_loss: 0.7016 - val_accuracy: 0.7457 Epoch 6/10 42/42 [==============================] - 6s 133ms/step - loss: 0.4148 - accuracy: 0.8378 - val_loss: 0.8888 - val_accuracy: 0.7155 Epoch 7/10 42/42 [==============================] - 6s 135ms/step - loss: 0.3828 - accuracy: 0.8462 - val_loss: 0.6643 - val_accuracy: 0.7931 Epoch 8/10 42/42 [==============================] - 5s 123ms/step - loss: 0.2999 - accuracy: 0.8919 - val_loss: 0.3769 - val_accuracy: 0.8664 Epoch 9/10 42/42 [==============================] - 5s 125ms/step - loss: 0.2474 - accuracy: 0.9177 - val_loss: 0.3600 - val_accuracy: 0.9009 Epoch 10/10 42/42 [==============================] - 5s 130ms/step - loss: 0.1429 - accuracy: 0.9528 - val_loss: 0.2745 - val_accuracy: 0.8966
Save the model
Loss and Accuracy of the model
output:
18/18 - 1s - loss: 0.1902 - accuracy: 0.9304 Restored model, accuracy: 93.04%
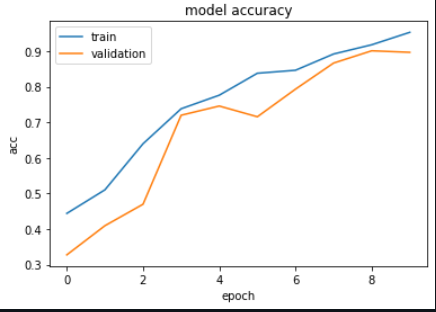
The Metrics
output:
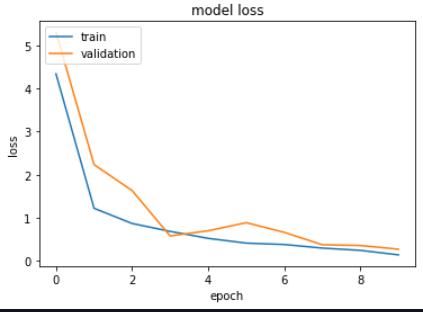
output:
Custom Predictions
output:

output:
\'Healthy\'
Cainvas Gallery: https://cainvas.ai-tech.systems/gallery/
Conclusion
We’ve trained our Deep Learning model using TensorFlow and Keras for detecting grape leaf disease and got an accuracy of 93%.
Notebook link is here
Credit: Om Chaithanya V
You may also be interested in
- Learning more about Fruits Classification using Deep Learning
- Reading about Mushroom Classification Using Deep Learning
- Also Read: Predicting Churn for Bank’s Customer using ANN
- Finding out about Computer Vision, the technology enabling machines to interpret complex data and mimic human cognition
Become a Contributor: Write for AITS Publication Today! We’ll be happy to publish your latest article on data science, artificial intelligence, machine learning, deep learning, and other technology topics
