Diabetes is a serious problem which many people face nowadays and which can lead to other serious health diseases. During the period of Covid-19 we also came to know that the conditions of a diabetic patient is much more critical than a non-diabetic patient.
So if we can take help of deep learning to predict the risk of getting diabetic and early prediction of diabetic will help people take care of their health and prevent themselves from getting diabetic.
Table of Content
- Introduction to cAInvas
- Importing the Dataset
- Data Analysis and Data Cleaning
- Trainset-TestSet Creation
- Model Architecture and Model Training
- Introduction to DeepC
- Compilation with DeepC
Introduction to cAInvas
cAInvas is an integrated development platform to create intelligent edge devices. Not only we can train our deep learning model using Tensorflow, Keras or Pytorch, we can also compile our model with its edge compiler called DeepC to deploy our working model on edge devices for production.
The Diabetes Prediction model which we are going to talk about, is also developed on cAInvas. All the dependencies which you will be needing for this project are also pre-installed.
cAInvas also offers various other deep learning notebooks in its gallery which one can use for reference or to gain insight about deep learning. It also has GPU support and which makes it the best in its kind.
Importing the dataset
While working on cAInvas one of its key features is UseCases Gallary. When working on any of its UseCases you don’t have to look for data manually. The data is in the of table and is present as csv format. We will load the dataset through pandas as a dataframe in our workspace.
Data Analysis and Data Cleaning
To gain information about the data that we are dealing with, we will use the command df.info() and we get the following information.
RangeIndex: 2000 entries, 0 to 1999 Data columns (total 9 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 Pregnancies 2000 non-null int64 1 Glucose 2000 non-null int64 2 BloodPressure 2000 non-null int64 3 SkinThickness 2000 non-null int64 4 Insulin 2000 non-null int64 5 BMI 2000 non-null float64 6 DiabetesPedigreeFunction 2000 non-null float64 7 Age 2000 non-null int64 8 Outcome 2000 non-null int64 dtypes: float64(2), int64(7)
Next we will remove the duplicate data, replace zero with NaN, then drop all NaN values from dataframe and again obtain the information. For this we can run the following commands;
TrainSet-TestSet Creation
Next step is to create the train data and test data. For this we will drop the ‘outcome’ column from the dataframe and store it in a variable. For the labels we will use the ‘outcome’ column and store it in another variable. Then we will use the scikit learn’s train-test split module to create the train and test dataset.
Model Architecture and Model Training
After creating the dataset next step is to pass our training data for our Deep Learning model to learn to classify Diabetic and Non-Diabetic Patients. The model architecture used was:
Model: "sequential" _________________________________________________________________ Layer (type) Output Shape Param # ================================================================= dense (Dense) (None, 8) 72 _________________________________________________________________ dense_1 (Dense) (None, 8) 72 _________________________________________________________________ dense_2 (Dense) (None, 4) 36 _________________________________________________________________ dense_3 (Dense) (None, 1) 5 ================================================================= Total params: 185 Trainable params: 185 Non-trainable params: 0 _________________________________________________________________
The loss function used was “binary_crossentropy” and optimizer used was “Adam”.For training the model we used Keras API with tensorflow at backend. The model showed good performance achieving a decent accuracy. Here are the training plots for the model:
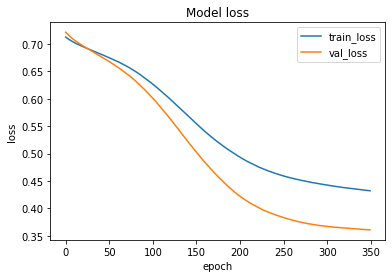
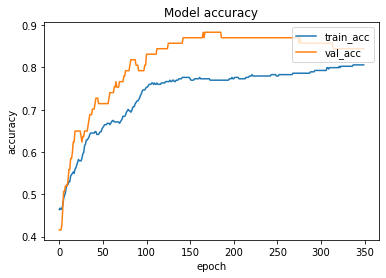
Introduction to DeepC
DeepC Compiler and inference framework is designed to enable and perform deep learning neural networks by focussing on features of small form-factor devices like micro-controllers, eFPGAs, cpus and other embedded devices like raspberry-pi, odroid, arduino, SparkFun Edge, risc-V, mobile phones, x86 and arm laptops among others.
DeepC also offers ahead of time compiler producing optimized executable based on LLVM compiler tool chain specialized for deep neural networks with ONNX as front end.
Compilation with DeepC
While training the model, we saved the best model using the Keras’ Model Checkpoint and saved it in H5 format using Keras as it easily stores the weights and model configuration in a single file.
After saving the file in H5 format we can easily compile our model using DeepC compiler which comes as a part of cAInvas platform so that it converts our saved model to a format which can be easily deployed to edge devices. And all this can be done very easily using a simple command.
And that’s it, it is that easy to create a Diabetes Prediction model which is also ready for deployment on edge devices.
Link for the cAInvas Notebook: https://cainvas.ai-tech.systems/use-cases/diabetes-prediction-app/
Credit: Ashish Arya
Also Read: Hate Speech and Offensive Language Detection
